Research
Overview
The Mathematics in Medicine Program at Houston Methodist focuses primarily on developing practical mathematical models and tools that can be used in the clinic by physicians to predict treatment outcomes for each individual patient prior to actual treatment. Our research involves multiple fields, including medicine, biology, engineering, mathematics, and physics. These tools are derived from fundamental principles of physics and biology, so they can be broadly applicable to many different fields of clinical sciences, including cancer, cardiovascular diseases, and infectious diseases, among others. More importantly, information to build and use these tools can be directly assessed from CT scans, patient tissue analyses, MRI, mammography, and other non-invasive or minimally-invasive procedures that the patient would normally receive within the standard of care. With these quantitative tools, frontline clinicians will be able to determine more effective, patient-specific drug treatment strategies (i.e., individualized medicine), such as the amount, frequency, and delivery platform and the need for ancillary non–drug-based treatments.
Mechanistic Modeling of Cancer Treatment
Biophysical theories to distinguish PDAC tumors with different pathological characteristics with Eugene J Koay, MD, PhD, Anirban Maitra, MBBS
Pancreatic ductal adenocarcinoma (PDAC) is generally associated with early distant metastasis and resistance to chemotherapy and radiation, and it lacks biomarkers to stratify patients based on disease severity and treatment needs. To this end, we are investigating computed tomography (CT) features of PDAC for their predictive and prognostic significance. We have identified a morphological imaging feature based on baseline standard of care CT scans that is associated with cellular, pathological, biological, and clinical characteristics of PDAC. This imaging feature can successfully separate patients into distinct prognostic groups and may be incorporated into clinical trials to personalize and improve the care of patients with this lethal disease. These results can be used in everyday standard clinical care as well as translational studies to identify how cancer-host interactions may be regulated and eventually manipulated to alter the natural course of disease.
Mathematical modeling of immunotherapy identifies novel early-time predictor of patient response with Eugene J Koay, MD, PhD, James Welsh, MD, David S Hong, MD
Working as part of an interdisciplinary team of engineers, physicians, and pathologists, we have developed a series of mathematical modeling descriptions of tumor-immunotherapy interactions and the associated patient therapeutic response. These models are built upon several key mechanistic factors involved in tumor immunotherapy response. Through careful planning, we have arrived at a final form of the model which includes only the most relevant factors in the immunotherapy-mediated immune response, allowing us to obtain unique quantification of these parameters by fitting the model to measured patient data. This effort has facilitated new insights into the most important biological parameters involved in successful immunotherapy treatment, providing valuable information about how immunotherapy may be modified to achieve greater treatment efficacy on a per-patient basis. We have also discovered a novel method for early-time clinical prediction of patient response to immunotherapy (that is, will the treatment be effective for the individual patient?), as well as a predictor of overall patient survival. There are several papers in review and in preparation which detail these exciting results.
Mathematical modeling of tumor response to drug treatment using non-coding RNAs with George A Calin, PhD, Bulent Ozpolat, PhD, Gabriel Lopez-Berestein, MD
Interest in the development of non-coding RNA-based therapeutics in cancer treatment has significantly increased in parallel with our understanding of the widespread role of non-coding RNAs in disease pathogenesis. It has been shown that treatment with non-coding RNAs could potentially reduce tumor growth and resensitize cancer to standard of care chemotherapeutics subsequent to development of drug resistance in vivo, without further identified toxicity. We are developing an integrated experimental and multiscale modeling approach to assist in understanding the effects of a variety of targeted, non-coding RNA-based nanotherapeutics on drug resistance in many different cancer systems. We expect to provide quantitative understanding into the efficacy and potential toxicity of treating these cancers by directly targeting non-coding RNAs in the tumor and its microenvironment, both alone and in combination with existing chemotherapeutics.
Development of a nomogram for predicting tumor regression outcome with neoadjuvant chemotherapy in CRCLM with Jason Fleming, MD, Daniel A. Anaya, MD
Surgical resection is the most effective treatment for colorectal cancer liver metastases (CRCLM), but the majority of tumors are not suitable for upfront surgery. A combination of the chemotherapy drugs neoadjuvant leucovorin, 5-fluorouracil, and oxaliplatin (FOLFOX), has become the recommended standard of care for treatment prior to surgery. We are investigating the variability in patient response to FOLFOX using pharmacokinetic and statistical modeling, and have identified patient-specific parameters that are the key determinants of response to chemotherapy. We are developing and validating a nomogram to provide a clinically useful tool to predict tumor outcome following neoadjuvant FOLFOX in CRCLM, which will offer clinicians a tool to decide a patient-specific therapy regimen to improve outcome.
Mathematical modeling of radiofrequency energy absorption in biological tissues with Steven Curley, MD
Non-invasive radiofrequency hyperthermia (NiRFH) is a clinical intervention that involves the application of high frequency electromagnetic radiations to produce mild hyperthermia in malignant tumors. While it has been extensively investigated clinically for several cancer types (e.g. central nervous system, lungs, breast), clinical studies of NiRFH for hepatic malignancies remain elusive. This is primarily due to technical challenges associated with the anatomic location of the liver and the high energy cost of transmitting the electromagnetic field through adipose tissue, which often leads to insufficient heating of the liver, overheating of subcutaneous fat and chest wall structures, and skin burns. We are working to develop a mechanistic model based on the first principles of physics to improve treatment efficacy of NiRFH on hepatic malignancies. Recent model results show that currently available NiRFH field equipment is inadequate to heat tumor tissues in the liver in a reliable and reproducible way. The model further suggests the application of a large number of multiphase array variable frequency RF beams may be necessary to attain the desired temperature rise in hepatic malignancies.
Imaging-based pharmacokinetic analysis of nanoparticles with C. Jeffrey Brinker, PhD
The advent of nanotechnology in medicine permits the packaging of medicinal cargo (drugs, diagnostic agents, biologics, etc.) inside nanocarriers, like nanoparticles and nanotubes, to deliver the cargo in a targeted fashion in the body. This is especially advantageous for cargo which is toxic, unstable, or insoluble. By isolating the cargo from the whole-body system and delivering it only at specific locations, nanomedicine has the potential to improve delivery efficiency and safety of the cargo. It has been recognized that the physicochemical characteristics of nanoparticles, like size, charge, and surface chemistry, play a critical role in governing the distribution of nanoparticles inside the body and affect their clearance from the body. To establish the quantitative relationship between particle characteristics and their biological behavior, we integrate longitudinal SPECT/CT imaging in rats with semi-mechanistic mathematical modeling. This provides information critical to making recommendations for nanoparticle design aimed at improving the distribution and clearance characteristics of nanoparticles for biomedical applications.
A modeling approach to predict breast cancer patient response to neoadjuvant chemotherapy with Renata Pasqualini, PhD, Wadih Arap, MD, PhD(c), Jeffrey Brinker, PhD
In treating breast cancer, treatment selection based on predictive biomarkers remains a significant challenge in disease management. We are developing a mathematical tool to help predict the likelihood of response to neoadjuvant chemotherapy using patient specific tumor vasculature biomarkers. A semi-automated, computer-aided analysis platform has been implemented to allow for increased measurement accuracy and rapid throughput in rendering model predictions. With this platform, we are evaluating a prospective and a retrospective cohort of patients undergoing neoadjuvant chemotherapy by collecting and analyzing clinically relevant data, including pre- and post-treatment pathology specimens and dynamic contrast-enhanced magnetic resonance imaging. Through this collaborative effort, we have correlated response to neoadjuvant chemotherapy with both pretreatment tumor vasculature biomarkers and model parameters. With this predictive tool, it is feasible to evaluate primary breast tumor vasculature biomarkers and predict response to treatment prior to treatment initiation in a patient specific manner, thereby allowing a precision approach to breast cancer treatment.
Multiscale Modeling of Cancer and Other Biological Systems
Understanding ductal carcinoma in situ (DCIS) invasion with Mary Edgerton, MD, PhD, Alastair Thompson, MD
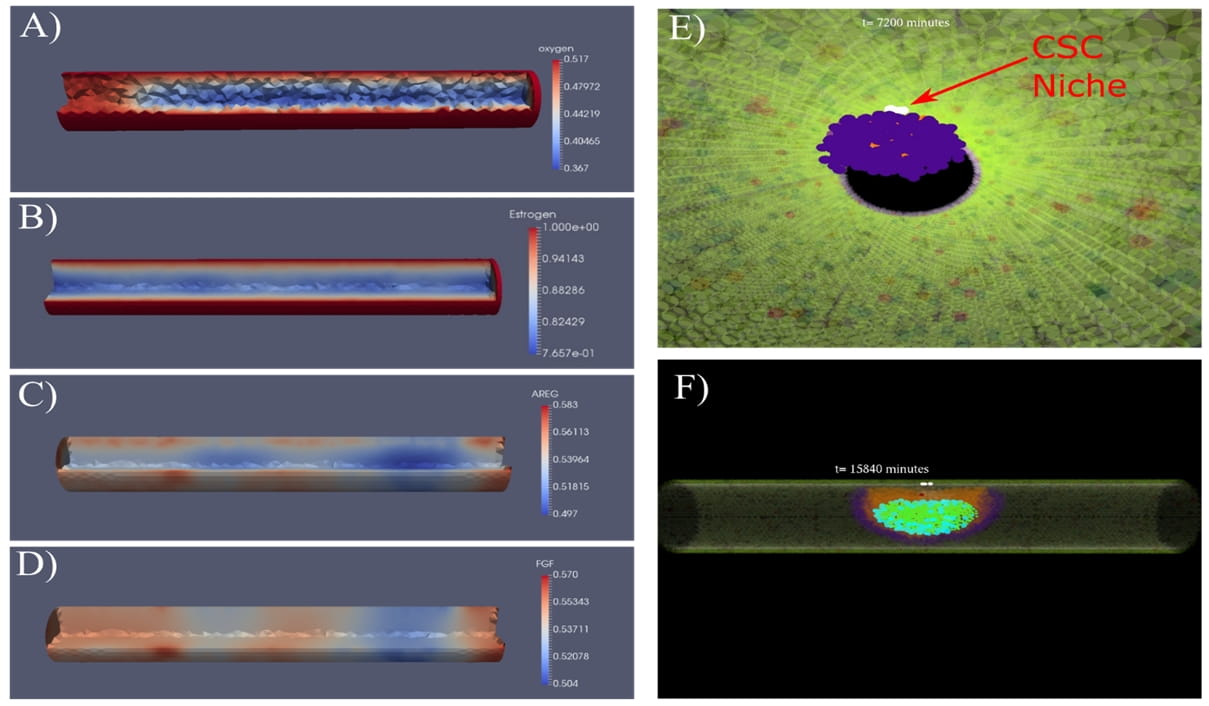
We are using multiscale agent-based modeling to study ductal carcinoma in situ (DCIS), the most common type of non-invasive breast cancer. By combining a discrete agent-based description of each individual cell within the tumor with a continuum mathematical description of molecules of interest within the mammary duct microenvironment (such as oxygen, estrogen, and some important downstream cell-cell signaling mechanisms), we are able to gain unique insight into the cell-scale phenomena involved in the earliest stages of DCIS progression. This work has led to notable insights into the molecular signaling control mechanisms involved in DCIS, and how they affect disease advance rates and overall phenotypic distribution within the cancer. We are currently using the model to gain new insight into how clinicians may achieve better treatment results, including optimizing surgical margins and the potential for directing the DCIS population phenotypic distribution to have more favorable response to cancer drug treatment.
Figure: Continuum (A-D) and discrete (E, F) scales within the DCIS model. Molecular profiles for all molecules of interest are solved mathematically as a continuum across the entire region of simulated duct, shown here for (A) oxygen, (B) estrogen, (C) amphiregulin (AREG), and (D) fibroblast growth factor (FGF). Cells in the model are represented discretely, as shown in panels (E) axial view from inside the mammary duct, and (F) side view from outside the duct, with mature duct wall shown as transparent so DCIS inside the duct is visible. DCIS is initiated through the transition of one or more cells in the mature duct wall to a tumor initiating cell phenotype (TIC), shown here as white cells in panel (D), and grows outward down the duct in both directions as regulated by cell rules and molecular signaling.
Upscaling and downscaling methods for functionally linking biological behaviors at different scales with John Lowengrub, PhD, Eugene J Koay, MD, PhD Anirban Maitra, MBBS
Due to the multiscale nature of biological systems, the underlying physiological processes that occur at different spatial and time scales should be quantified and linked together in a consistent manner. This presents an opportunity to push the boundaries of knowledge in current biological and biomedical investigations, using multiscale modeling as a platform for hypothesis generation and testing. We are developing a new class of multiscale methods that use directly measurable quantities at the cell scale to inform the model parameters at the tissue scale through rigorous upscaling techniques to close the continuum equations at the tissue scale. We are currently applying these new multiscale methods to estimation of pancreatic neoplasms to invasive pancreatic ductal adenocarcinoma using a large-scale human patient dataset of pancreas cystic lesions obtained from our clinical collaborators. The new tools developed here have potential to impact not only tumor biology, but other multiscale problems in the biological sciences including development, wound healing, tissue regeneration, tissue-based epidemiology and ecology.
Normal mammary gland development with Michael T Lewis, PhD, and Jeffrey M Rosen, PhD
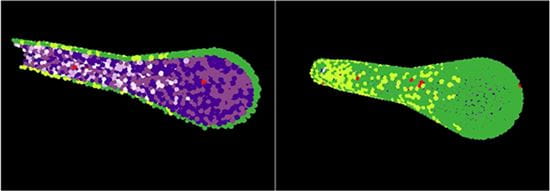
We are developing a multiscale, agent-based model of the mammary terminal end bud (TEB) to study the cellular and molecular mechanisms involved in pubertal development of the mammary gland. The model includes both discrete models of cells, which are represented uniquely on a cell-by-cell basis, as well as several of the key molecular signaling pathways involved in organ development, including estrogen and the downstream epithelial to stromal AREG to FGF pathway. Through explicit linking of the molecular and cell scales, we are able to study how perturbations in molecular signaling, cell types, and cell decision-making rules affect overall gland development. This model allows us to achieve mathematical quantification of biological parameters that may not be available in the literature to successfully reproduce observed biological behavior, and also provides a jumping-off point for studies of the ways normal biological systems may lead to disease when they fail to function properly.
Figure: 3D TEB shown in both cross-section (left) and in full (right). TEB is composed of outer myoepithelial (green) and inner luminal (purples) layers, and cells are approximated mathematically as spherical bodies which grow, divide, and participate in and respond to molecular signaling.