- Home
- Medical Research
- Cancer Research
- Chao Center for Brain Health Research
- Home
- Medical Research
- Cancer Research
- Chao Center for Brain Health Research

Ting Tsung and Wei Fong Chao Center for BRAIN
Ting Tsung and Wei Fong Chao Center for BRAIN
The Chao BRAIN Center operates five research programs, including: (1) Alzheimer's Disease/Parkinson's Disease (AD/PD) Mechanisms and Therapeutics, (2) Stroke Detection and Diagnosis, (3) Brain Tumor Mechanisms and Therapeutics, (4) Neuronal Imaging, (5) Digital NeuroHealth.
Our Research Programs
-
ADPD Mechanisms and Therapeutics
Project: Systematic Alzheimer's Disease Drug ReposiTioning (SMART) framework Based on Bioinformatics-guided Phenotype Screening and Imagomics Modeling
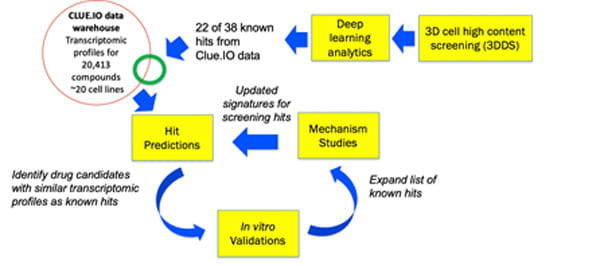
The SMART framework for drug repositioning and discovery.
Combining the systems pharmacology and drug repositioning expertise of the Wong Lab at the Houston Methodist with the Alzheimer’s biology expertise of the Kim and Tanzi labs at Massachusetts General Hospital, we propose a SysteMatic Alzheimer’s disease drug ReposiTioning (SMART) framework based on bioinformatics-guided phenotype screening. Reformatting a novel three-dimensional human neural stem cell culture model of AD (a.k.a. Alzheimer's in a dish) developed in the Kim and Tanzi labs for high content screening, the Wong lab screened 2,640 known drugs and bioactive compounds and obtained a panel of 38 primary hits that strongly inhibit β-amyloid-driven p-tau accumulation. We hypothesize that iteratively running relatively small screens with our novel 3D cell model and applying artificial intelligence (AI) modeling to the transcriptomic profiles of the screening hits will allow us to quickly obtain a panel of robust novel drug candidates for AD and gain an in-depth understanding of disease mechanisms from those repositioned drug candidates, which will subsequently improve the success rate of predicting novel hits. Success of this work will lead to new AD therapeutic compounds ready for translation into clinical trials and a deeper understanding of molecular mechanisms of AD pathophysiology. In addition, the SMART framework can be generalizable to other big data driven drug discovery studies.
Project: Brain Microenvironment Crosstalk Modeling and Combination Drug Repositioning
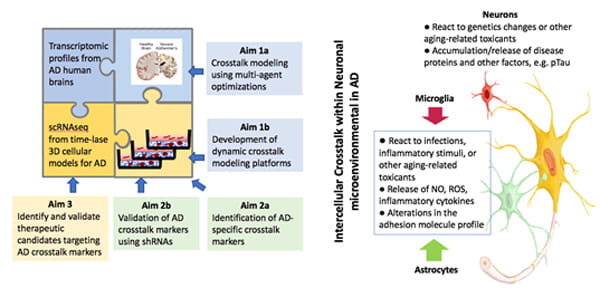
Identification of novel AD-specific crosstalk pathways among neurons and glial cells using the SCRBI explorer and 3D human cell culture models of AD.
There is mounting evidence that non-neuronal brain cells including astrocytes, oligodendrocytes, and microglia, play crucial roles in Alzheimer's disease pathogenesis by interacting with neurons in various stages of the disease. Understanding altered neuron-glia and glia-glia crosstalk in AD is essential to comprehensively understand pathogenic cascade of AD, which may hold the key for identifying therapeutic targets. Recent progresses in "single-cell biology" makes it possible explore pathological pathways of AD at the cellular level. These unbiased, cell-type specific datasets provide a new insight how different brain cells and their interactions contribute to AD pathology.
This project aims to develop a crosstalk modeling tool and human cellular models to identify altered neuron-glia and glia-glia crosstalk pathways in AD. We believe that the combination of multi-cellular systems biology modeling and novel AD models will identify AD-specific neuron-glia and glia-glia crosstalk pathways and novel drug targets. We are developing a multicellular systems biology modeling tool, Single-Cell Resolution Brain Interactome (SCRBI) Explorer to investigate intercellular crosstalk networks among different subgroups of neurons and glial cells and analyze cell-type and subgroup omics data to predict crosstalk pathways in a wholistic and unbiased fashion. Identified pathways and ligand and receptor pairs will be evaluated for their therapeutic potentials. The success of this study will provide a powerful multi-cellular systems biology tool to unbiasedly model and identify neuron-glia and glia-glia crosstalk pathways and novel targets for AD therapeutics.
Project: Identification of Mechanisms and Potential Therapeutic Substrates for AD via Systems Neurobiology Approaches
Development of new cellular models of AD/PD and conducting high throughput screening FDA-approved drugs on imaging-based cell assays and preclinical studies to evaluate the translational potential of newly-identified repositioned drug candidates. Neuroscience and bioinformatics tools are deployed and integrated to study AD&PD at molecular, cellular, organ, and whole system levels on multiple platforms (in silico, in vitro, and in vivo). Drug efficacy, PK/PD, and chronic toxicity are examined in several transgenic mouse models.
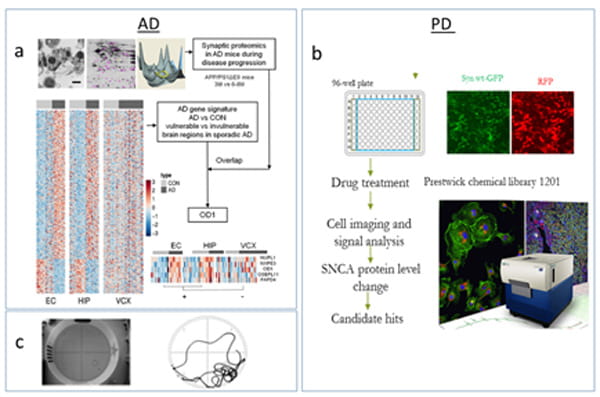
This project aims to identify novel mechanisms underlying the complex etiology of AD and potential therapeutic substrates, focusing on key substrates contributing the neurodegeneration or mediating the complex interaction between AD pathological events. Understanding the molecular and cellular mechanisms for the neurodegeneration at early disease stages of Alzheimer's disease will help identify potential effective therapeutic targets for this devastating disease. Hyperamyloidosis in the brain is known as the earliest neuropathological change and a unique etiological factor in AD, while progressive neurodegeneration in certain vulnerable brain regions forms the basis of clinical syndromes. It is not clear how early hyperamyloidosis is implicated in progressive neurodegeneration and what factors contribute to the selective brain vulnerability in AD. We have developed a workflow by integrating computational and experimental neurobiology methods to identify novel factors involved in the early neurodegeneration and brain vulnerability in AD. We examine neurodegeneration-specific gene signatures from sporadic AD patients and synaptic protein changes in transgenic AD mice. Subsequentially, we will assess the association of a top candidate gene with AD and investigate its mechanistic role in neurodegeneration.
Project: In vitro Drug Screening and in vivo Validation of Repositioned Drugs and Traditional Chinese Medicine Compounds for AD/PD Therapeutics
In this project, we are screening known drugs or bioactive compounds as well as herbal compounds of traditional chinese medicine (TCM) on human-derived cellular models of AD/PD and identifying drug hits via systems biology methods for evaluation on transgenic animal models. Disturbed proteostasis is known as a key event contributing to cell death in a variety of neurodegenerative diseases. Abeta/tau aggregates in AD and alpha-synuclein accumulation in PD can be reproduced in human cells (i.e. iPSC-derived neurons) while promoting the clearance of aggregated tau or alpha-synuclein has the potential to translate into therapeutics. Since the development of new therapies is a risky, costly and prolonged process, repurposing known drugs or bioactive compounds is considered an attractive strategy for drug development. Promising repositioning drug hits will be tested on mice bearing disease-relevant mutant before clinical trials. We perform automated drug screening using FDA-approved drug libraries and TCM herbal compounds to identify drug hits reducing toxically modified tau proteins or wild type of alpha-synuclein. We standardized a pipeline for preclinical drug testing in small animals and set up an animal behavioral suite for phenotype examination via Morris water maze, fear conditioning system, locomotion system, and Rotarod.
-
Stroke Detection and Diagnosis
Project: Stroke Lesion MRI Segmentation for Patient Outcome Prediction
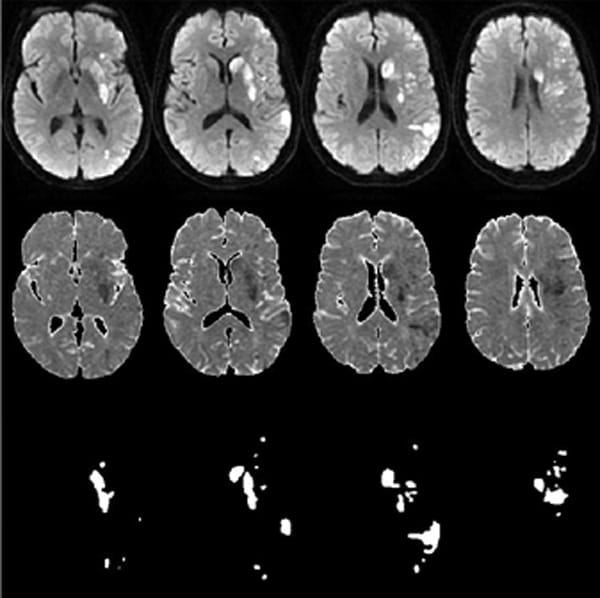
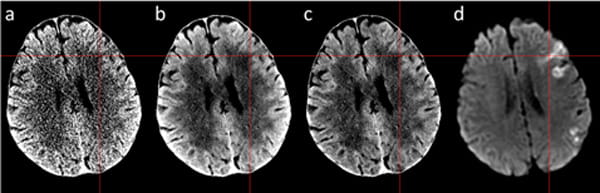
Representative stroke infarct segmentation using diffusion MRI (top row), apparent diffusion coefficient map (middle row) as input to AI model and the segmented results (bottom row).
Infarct volume is an independent predictor of 90-day modified Rankin Score (mRS) outcome in acute ischemic stroke (AIS). Deep learning image segmentation of infarct volume has shown promising results recently. Unfortunately, these all are based on traditional convolutional neural networks and can generate unpredictable results when input images are mirrored, flipped, or rotated. Manual measurement is laborious, and its variance is hard to quantify. This research aims to develop automated segmentation methods with built-in robustness for image rotation and mirror flipping. We have validated the accuracy over a large range of lesion sizes in more than 1,000 stroke patients, including a cohort of 175 cases who underwent thrombectomy. The automated method can segment acute stroke volume, even for infarct in choroid plexus. Segmented acute stroke volume is a significant prognostic indicator of 90-days outcome measured by mRS even after adjusting the effects of patients age and baseline NIH stroke scale (NIHSS).
Project: Deep Denoising of non-contrast Brain CT Scans
(a) Original NCCT, (b) denoised CT, (c) bias field corrected denoised CT, (d) diffusion MRI. The red crosshair corresponds to acute ischemic stroke location that has been irreversibly damaged.
In acute ischemic stroke, tissue ischemia caused an increase of tissue water. During the first 3 hours of acute ischemic stroke, we typically see 2-4 Hounsfield Units decrease in the stroke region on non-contrast CT (NCCT) images. Compared to image noise normally around 4-7 Hounsfield Units at full-dose radiation, early acute stroke detection thus presents a challenge for disease diagnosis and interpretation. This research project investigates a novel CT enhancement and noise reduction method using advanced deep learning algorithms to boost the image signal-to-noise (SNR) level comparable to high-quality MRI for improved neurological disease detection. In addition, the proposed deep denoising method is based on image-space and can be applied to any non-contrast head CT scans from varying vendors to improve disease detection. It is especially useful in emergency settings when MRI is not available. A prospective clinical trial on the technology in acute ischemic patients is ongoing at Houston Methodist.
Project: Multimedia Intelligence for Stroke Screening and Assessment
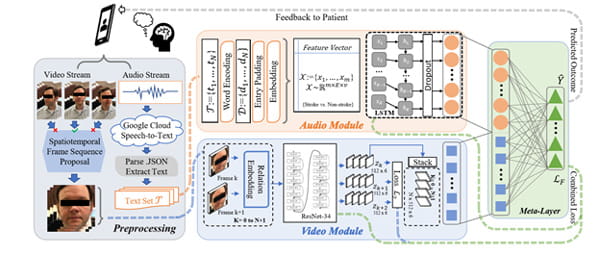
Dataflow diagram of the video/audio stream network fusion process for stroke/TIA prediction using the feature output from the ResNet-34 before the final fully-connected layer.
Acute ischemic stroke is a common but challenging disease to diagnose in the emergency setting. Facial asymmetry is a frequent finding in patients with stroke. This work proposes an AI framework for screening and assessing facial asymmetry caused by stroke using video analysis on patient videos from an emergency room setting, performing spatiotemporal analysis on facial asymmetry and providing a clinical decision tool to support emergency diagnosis. Clinical datasets for this study were acquired by the Eddy Scurlock Comprehensive Stroke Center at Houston Methodist with susceptible stroke patients and healthy volunteers as controls. Subjects are asked to first say the phrase, “Today is a sunny day in Houston, Texas,” and describe a picture known as the “Cookie Theft” picture while being recorded. Videos are labeled with the clinical impressions on whether or not patients have a stroke or transient ischemic attack (TIA), as well as discharge diagnosis for validation. Acquisition of facial data in natural settings makes our work more robust and useful for clinical use.
-
Brain Tumor Mechanisms and Therapeutics
Project: Chronic Stress and Brain Metastasis from Triple-negative Breast Cancer
Brain metastatic tumor cells have intrinsic or are able to acquire brain-like characteristics to assimilate into the neural network. Especially neurotransmitters, gliotransmitters, neurotrophic factors, and neural cytokines, called neuroactive substances (NSs) are the most abundant and key intercellular communication signals in the brain. Tumor cells are most likely to leverage them to favor their outgrowth in the brain. Chronic stress is often accompanied by aberrant or imbalanced levels of brain NSs and is prevalently (50-75%) experienced in cancer patients. Epidemiological and clinical studies provided strong evidence for links between chronic stress and metastasis, especially in younger female patients. Triple negative breast cancer occurs most frequently in younger women with a higher prevalence of abdominal obesity and metabolic syndrome linked to vulnerable neuroendocrine regulations to stress. Clinically, intake of beta-blocker drugs, in reducing stress-related signals, is associated with improved metastasis-free survival in TNBC patients. We have showed that chronic stress promotes the development of brain metastasis from TNBC and thus hypothesize that a chronic stress-induced brain metastatic niche or even a pre-metastatic niche with aberrant NSs creates a favorable or receptive microenvironment for TNBC brain metastasis. Our goal is to study how TNBC cells and various neural cells co-opt chronic stress-altered NSs in the metastatic and pre-metastatic niches in the brain and identify effective therapeutic or preventive strategies for TNBC brain metastasis.
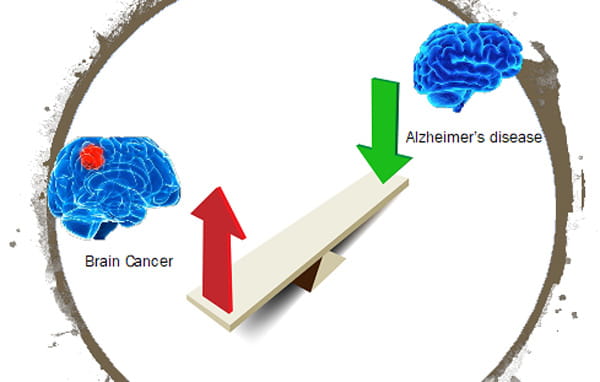
Project: Brain Cancer excludes Alzheimer's
In this project, we investigate how the presence of tumor cells in the brain would reduce pathogenesis of Alzheimer’s disease (AD). Both AD and brain cancer increase with age. However, several comprehensive epidemiologic studies with more than one million participants have reported a consistent inverse association between the two diseases, and the risk of AD in individuals with a history of cancer was decreased up to 35%. In addition, many factors upregulated in cancer to sustain growth and cell survivals are founded downregulated in AD, contributing to neurodegeneration. Since astrocytes are important in AD pathogenesis through interactions with adjacent neurons, microglia, and oligodendrocytes as well as secretion of significant quantities of Aβ to increase amyloid burden in the brain, we hypothesize that the tumor-astrocyte interactions suppress some astrocyte-initiated AD pathogenesis pathways, including reducing Aβ production from neurons and astrocytes, facilitating Aβ break-down by astrocytes, and regulating neuroinflammation. In this study, we will apply novel system biology modeling and multi-omics technology to investigate how astrocyte-tumor crosstalk regulates AD pathogenesis and identify novel crosstalk target for developing new therapeutic strategies for combating AD.
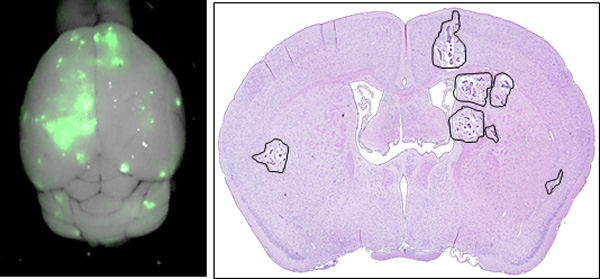
Project: Systematic Drug Repositioning for Medulloblastoma
Medulloblastoma (MB) is the most common malignant brain tumor of childhood. Although outcomes have improved in recent decades, new treatments are still needed to improve survival and reduce treatment-related complications. MB sub-types Groups 3 and 4 represent a particular challenge due to their intra-group heterogeneity, which limits the options for “rational” targeted therapies. We developed a systematic drug repositioning platform that integrates a non-parametric, bootstrapping-based simulated annealing algorithm and a 3D functional drug network to characterize dysregulated driver signaling networks, thereby identifying potential drug candidates. From over 1,300 drug candidates studied, we identified five members of the cardiac glycoside family as potentially inhibiting the growth of Groups 3 and 4 MB, and subsequently confirmed this in vitro. Systemic in vivo treatment of patient-derived orthotopic xenograft (orthotopic PDX) models of Groups 3 and 4 medulloblastoma with digoxin, one of the five cardiac glycosides, prolonged animal survival at plasma levels known to be tolerated in humans. Furthermore, single-agent digoxin has efficacy comparable to 20 Gy of radiation. The treatment of mice with a combination of radiation therapy and digoxin resulted in a statistically significant improvement in survival over the radiation-only group. These results demonstrate the power of the systematic drug repositioning method in identifying a potential treatment for MB. Our strategy could potentially be used to accelerate the repositioning of treatments for other human cancers that lack clearly defined druggable targets.
Project: Prognostic Gene Discovery in Glioblastoma Patients using Deep Learning
This project aims to apply deep machine learning to identify specific novel genes that are potential biomarkers or therapeutic targets of glioblastoma (GBM). Conventional machine learning approaches have been used to determine the gene expressions that are prognostic to glioblastoma patient survival. These approaches often require dropping a large number of genes in order to fit the survival outcome, hindering biological pathway interpretation and introducing random bias toward the selected gene factors. Using 3,581 differentially expressed genes from the GBM-specific TCGA database as inputs, we developed a deep learning model to predict patient survival prognosis. The deep learning features are combined with age, gender, MGMT methylation, G-CIMP, IDH1/2 mutation and cancer subtype to an integrative prognostic model.
-
Neuronal Imaging
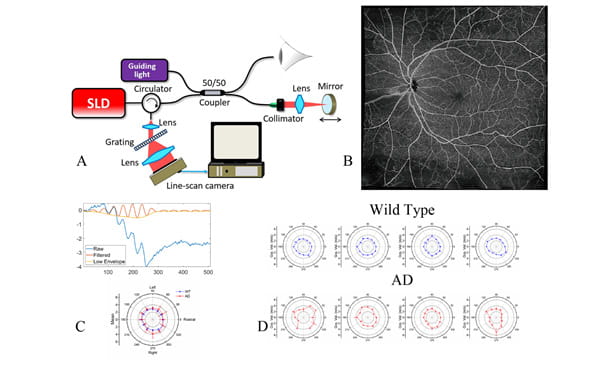
Project: Multi-modal optical coherence tomography probe for detection and monitoring of Alzheimer’s disease
(A) Schematic of the OCT system. (B) Blood distribution (OCT angiography image). (C) OCT. elastography (OCE) signal and result of obtaining the biomechanical properties of retina, respectively. (D) OCE results of retina from wild type and AD mouse model.
This research project aims to develop low-cost, non-invasive imaging methods to detect and monitor the progression of Alzheimer’s disease in patients. Studies have shown that the retina, a central nervous system tissue formed as a developmental outgrowth of the brain, is profoundly affected by AD. Many detectable disease-specific symptoms that can be used to detect AD in the early stage or to monitor the progress of AD development, such as substantial ganglion cell degeneration, thinning of the retinal nerve fiber layer, and loss of axonal projections in the optic nerve. These changes, as well as the known gathering of amyloid β-protein (Aβ) plaques, can further alter the biomechanical properties of retina. We designed and investigated a spectrum domain, portable, label free Optical coherence tomography (OCT) system to acquire real-time, high-resolution 3D retina images to identify and extract quantitative features of eye volumes, nerve and blood vessel distributions, and biomechanical properties of lens and retina in real time and then combine these features into training a deep learning model to detect and classify stages of the disease. It can serve a great tool to assess retinal abnormalities in vivo and provide multiple parameters for the distinction between normal aged individuals and patients with neurodegenerative diseases. Since OCT has been widely used in clinics to detect and monitor ophthalmology diseases, the detection and monitoring of neurodegenerative diseases can be performed during routine ophthalmology examinations.
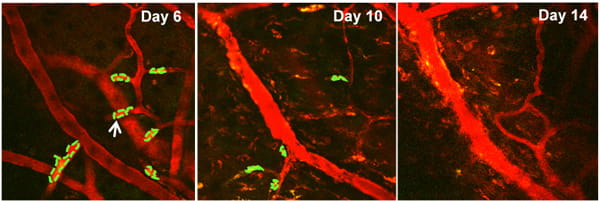
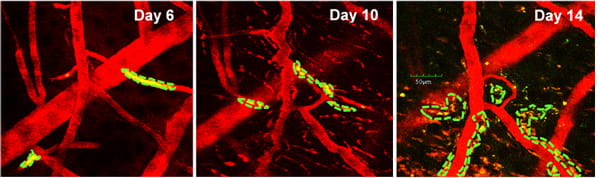
Project: Intravital multi-photon imaging for brain disorder studies
The intra-vital multi-photon microscopy at the TT & WF Chao Center for BRAIN allows in vivo, dynamic subcellular resolution imaging beyond the depth limits of conventional confocal microscopy. Using this in vivo imaging technique, various neural cells and brain tumor cells as well as their interaction could be visualized within the brain tissue of a living small animal model. An additional SIM scanner and various associated laser beams enable simultaneous multi-photon imaging with direct laser stimulation, allowing the researchers not only to view and capture remarkable 3-D images of tissues, but also to stimulate and excite specific cells or networks of associated cells in order to probe biological mechanisms and develop therapeutic strategies.
Details of related cellular and tissue imaging equipment are available here.
-
Digital Neuralhealth
Project: Readmission Risk Assessment of Mental Health Patients
In collaboration with the Psychiatry Department and Systems Quality Group of Houston Methodist, this research project aims to apply machine learning approach to identify psych patients who are at high-risk and lists personalized post-acute care intervention that is best suited for individual patients. Evidence suggests that mental health conditions and symptoms can increase physical health readmission rates. Depression is an independent risk factor for many comorbid conditions. Little is known about predicting psychiatric readmissions and identifying modifiable factors that may reduce early readmissions. The study would also help hospitals, insurers, and homecare agencies improve the quality of care for patients while reducing 30-day readmission rates. Readmission risk assessment tool will determine if interventions designed to treat mental health symptoms can effectively reduce readmission risk. While our focus is on the psychiatric conditions, we will also focus on adult patients hospitalized for any physical health condition to identify interventions that could be adapted for study in our populations of interest. Readmission Risk Assessment tool will be administered any time before discharge and stratify the patients into two groups of Low Risk and High Risk of readmission.
Project: Hospital Outcomes and Readmission Risk Assessment for Hospitalization in Geriatric Neurodegenerative Patients
More than 5 million Americans are living with Alzheimer’s disease, and at least 1 million Americans live with Parkinson's disease. The risk of being affected by a neurodegenerative disease increases dramatically with age. Geriatric patients will use up more healthcare costs and geriatric patients with neurodegenerative conditions stay in the hospitals more often than other people of the same age group because of their neurological deficits and functional decline. However, hospitalization may be unavoidable due to many reasons, such as trauma, infection, stroke, surgery, or exacerbation of systemic illnesses. Therefore, it is important to understand all the factors that are significant to lessen the hospital ordeal and to achieve a good outcome post hospitalization. In collaboration with the Department of Neurology and the Neuro ICU, the objective of this project is to develop machine learning models that can predict: (1) by 1st or 2nd day of admission, the outcomes of hospitalization of geriatric patients with neurodegenerative diseases, notably dementia, Alzheimer’s, and Parkinson’s, and (2) the risk of readmission of these patients in order to devise effective follow-up interventions for modifiable factors that could improve patients’ outcomes and reduce readmission risks. We are applying machine learning to analyze ten-year’s worth of dementia patient data at Houston Methodist and narrowing down risk factors (both modifiable and non-modifiable) vital in determining patient outcomes. Accurate prediction of readmissions in this patient segment could also help mitigate rising health costs.
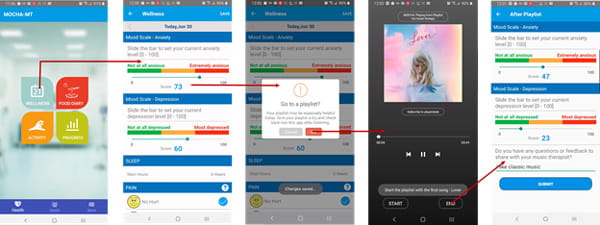
Project: Personalized Digital Music Therapeutics for Mental Health Patients
In collaboration with the Houston Methodist Center for Performing Arts & Medicine, this project aims to study the effects of music therapy (MT) and other musical interventions on different aspects of neurological disorders, including mood, anxiety, depressive syndromes, and quality of life on neurological patients. Psychological distress, depressive syndromes, and mood disorders represent typical comorbid conditions in neurological disorders that can lead to acute and chronic stress responses. Cognitive neuroscience research over the past decade has created increasingly fascinating insights into how the human brain processes music, while also enhancing the understanding of brain processes related to many mental health disorders. Music therapy is considered a complementary treatment that can be utilized in conjunction with psychiatry and psychology to treat and manage mental health needs. People with moderate symptoms of anxiety and depression may benefit from self-directed protocols for mood symptom management, particularly when paired with therapist-directed intervention and guidance. This project implements individual music therapy for mood symptom management through a blended model incorporating music therapist-directed and self-directed treatment components. MOCHA digital therapeutics, which has previously been tested successfully for cancer patients and survivors, encourages independent accountability and self-reinforcement in maintaining wellness plus a means of ongoing communication with healthcare providers, which may help to mitigate neurological disorders. A workflow of the patient-side app of music therapy delivery via MOCHA digital therapeutics is shown above.
Our Resources
The following list of resources is available to provide more information about the diseases and disorders studied and tools developed at the Chao Center for BRAIN:
- Alzheimer’s Association
- International Society to Advance Alzheimer’s Research and Treatment (ISTAART)
- Alzheimer Research Forum
- National Institute on Aging Alzheimer’s Disease Education and Referral (ADEAR) Center
- Cure Alzheimer’s Fund
- NIA Alzheimer’s Disease Research Centers
- Harvard Medical School Screening Facility
- Harvard NeuroDiscovery Center
Contact Information
Ting Tsung and Wei Fong Chao Center for BRAIN
Houston Methodist
6670 Bertner Ave., R6-South
Houston, TX 77030
713.441.3726
Clinical Trials
At Houston Methodist, our dedicated teams of world-renowned researchers help support the mission of our Ting Tsung and Wei Fong Chao Center for Brain of bringing the latest technologies and advanced treatment options to patients as quickly and safely as possible.